Genomic conservation of two species of the order Asparagales with bimodal karyotype, using genomic in situ hybridization (GISH)
DOI:
https://doi.org/10.18387/polibotanica.60.22Keywords:
Agave, Aloe, GISH, Bimodal KaryotypeAbstract
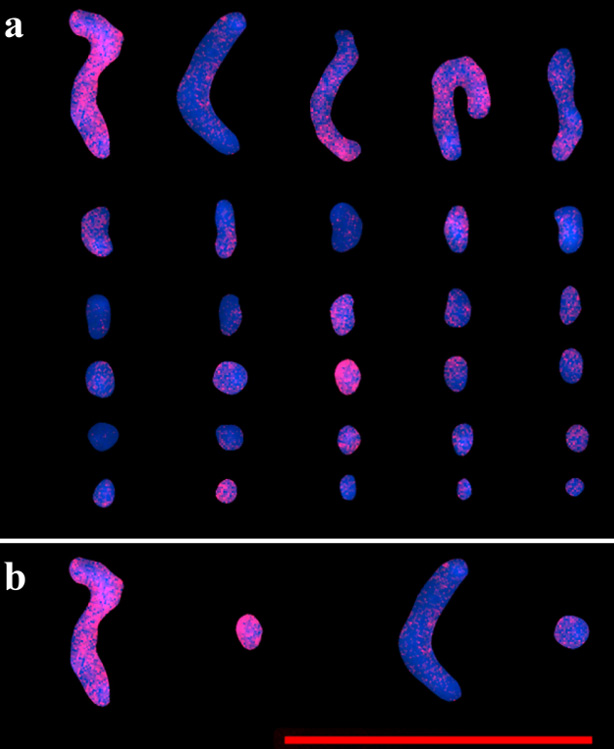
The Asparagales order, one of the main subdivisions of the monocots, comprises 14 families and more than 36,000 species, notable for its high rate of diversification. Within this order, a wide variability in genome size and karyotype is observed, with some genera showing a bimodal karyotype. The genus Aloe, belonging to the family Asphodelaceae, is notable for its diversity and although most are diploid, tetraploid and hexaploid species have also been identified. Despite this variability in chromosome number, Aloe's bimodal karyotype remains relatively stable, with differences in DNA content between species. In contrast, the Agave genus of the Asparagaceae family, presents a conserved bimodal karyotype with 30 chromosomes in the haploid genome, which includes 5 large chromosomes and 25 small chromosomes. Agave species show natural variations in ploidy, but the basic karyotype pattern is preserved. Phylogeny based on whole genomes suggests that Agave is closely related to other genera such as Manfreda and Polianthes. Research on in situ hybridization and the GISH technique has made it possible to identify and analyze genomic rearrangements and karyotype components in various species. In this context, a study has been carried out to hybridize the genome of Aloe vera in interphase nuclei and metaphase cells of hybrid Agave H11648 to evaluate how an evolutionarily distanced genus integrates into the Agave genome, seeking to better understand the karyotype dynamics in these plants. Our results showed preservation of genomic regions of Aloe in Agave by observing genomic hybridization in all chromosomes and in interface nuclei in hybrid Agave H11648. These data strongly suggest that the bimodal karyotype of agaves was not acquired by the fusion of genomes with different chromosome sizes, but rather by rearrangements, fusions, and fissions during the period of evolution of the genus.
References
Ali HBM, Lysak MA, Schubert I (2004) Genomic in situ hybridization in plants with small genomes is feasible and elucidates the chromosomal parentage in interspecific Arabidopsis hybrids. Genome 47 (5):954-960. doi:https://.doi.org/10.1139/g04-041
Baez M, Vaio M, Dreissig S, Schubert V, Houben A, Pedrosa-Harand A (2019) Together But Different: The Subgenomes of the Bimodal Eleutherine Karyotypes Are Differentially Organized. Front Plant Sci 10. doi:https://.doi.org/10.3389/fpls.2019.01170
Bogler DJ, Simpson BB (1996) Phylogeny of Agavaceae based on ITS rDNA sequence variation. Am J Bot 83 (9):1225-1235. doi:https://.doi.org/10.2307/2446206
Brandham PE, Doherty MJ (1998) Genome size variation in the Aloaceae, an angiosperm family displaying karyotypic orthoselection. Ann Bot-London 82:67-73. doi:https://.doi.org/10.1006/anbo.1998.0742
Carter S, (1994) Flora of Tropical East Africa - Aloaceae (1994). Royal Botanic Gardens K. Taylor & Francis.
Castorena-Sánchez I, Escobedo RM, Quiroz A (1991) New cytotaxonomical determinants recognized in six taxa of Agave in the sections Rigidae and Sisalanae. Canadian Journal of Botany 69 (6):1257-1264. doi:https://.doi.org/10.1139/b91-163
Chase MW, Christenhusz M, Fay M, Byng J, Judd WS, Soltis D, Mabberley D, Sennikov A, Soltis PS, Stevens PF (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical journal of the Linnean Society 181 (1):1-20. doi:https:// /10.1111/boj.12385
Chester M, Riley RK, Soltis PS, Soltis DE (2015) Patterns of chromosomal variation in natural populations of the neoallotetraploid Tragopogon mirus (Asteraceae). Heredity 114 (3):309-317. doi:https://.doi.org/10.1038/hdy.2014.101
Eguiarte LE, M. R. Duvall, G. H. Learn Jr, and M. T. Clegg (1994) The systematic status of the Agavaceae and Nolinaceae and related Asparagales in the Monocotyledons: An analysis based on the rbcL gene sequence. Botanical Sciences (54):35 - 56. doi:https://.doi.org/10.17129/botsci.1427
Elliott TA, Gregory TR (2015) What's in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philosophical Transactions of the Royal Society B: Biological Sciences 370 (1678):20140331. doi:https://.doi.org/10.1098/rstb.2014.0331
Gomez-Rodriguez VM, Rodriguez-Garay B, Palomino G, Martinez J, Barba-Gonzalez R (2013) Physical mapping of 5S and 18S ribosomal DNA in three species of Agave (Asparagales, Asparagaceae). Comp Cytogenet 7 (3):191-203. doi:10.3897/CompCytogen.v7i3.5337
Good-Avila SV, Souza V, Gaut BS, Eguiarte LE (2006) Timing and rate of speciation in Agave (Agavaceae). Proceedings of the National Academy of Sciences 103 (24):9124-9129. doi:https://.doi.org/10.1073/pnas.0603312103
Gunjan K, Roy BK (2010) Karyotype studies in dominant species of Aloe from eastern India. Caryologia 63 (1):41-49. doi:10.1080/00087114.2010.10589707
Halpin KM, Fishbein M (2013) A Chloroplast Phylogeny of Agavaceae subfamily Chlorogaloideae: Implications for the Tempo of Evolution on Serpentine Soils. Syst Bot 38 (4):996-1011. doi:10.1600/036364413x674850
Hasterok R, Ksiazczyk T, Wolny E, Maluszynska J (2005) FISH and GISH analysis of genomes. Acta Biol Cracov Bot 47 (1):185-192
Herrera JC, Romero JV, Camayo GC, Caetano CM, Cortina HA (2012) Evidence of Intergenomic Relationships in Triploid Hybrids of Coffee (Coffea sp.) as Revealed by Meiotic Behavior and Genomic in Situ Hybridization. Trop Plant Biol 5 (3):207-217. doi:https://.doi.org/10.1007/s12042-012-9105-x
Humphreys MW, Thomas HM, Morgan WG, Meredith MR, Harper JA, Thomas H, Zwierzykowski Z, Ghesquiere M (1995) Discriminating the Ancestral Progenitors of Hexaploid Festuca-Arundinacea Using Genomic in-Situ Hybridization. Heredity 75:171-174. doi:DOI 10.1038/hdy.1995.120
Jahan B, Vahidy AA, Saeed R, Mirbahar AA (2014) Karyological Studies in Ten Different Populations of Desert Lily Aloe Vera from Pakistan. Pak J Bot 46 (5):1731-1734
Kato A, Vega JM, Han F, Lamb JC, Birchler JA (2005) Advances in plant chromosome identification and cytogenetic techniques. Curr Opin Plant Biol 8 (2):148-154. doi:https://doi.org/10.1016/j.pbi.2005.01.014
Lee YS, Park HM, Kim NH, Waminal NE, Kim YJ, Lim KB, Baek JH, Kim HH, Yang TJ (2016) Phylogenetic relationship of 40 species of genus Aloe L. and the origin of an allodiploid species revealed by nucleotide sequence variation in chloroplast intergenic space and cytogenetic in situ hybridization. Genet Resour Crop Ev 63 (2):235-242. doi:https://.doi.org/10.1007/s10722-015-0243-5
Leitch IJ, Leitch AR (2013) Genome Size Diversity and Evolution in Land Plants. In: Greilhuber J, Dolezel J, Wendel JF (eds) Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Springer Vienna, Vienna, pp 307-322. doi:10.1007/978-3-7091-1160-4_19
Magallón S, Castillo A (2009) Angiosperm diversification through time. Am J Bot 96 (1):349-365. doi:10.3732/ajb.0800060
Markova M, Vyskot B (2009) New Horizons of Genomic in situ Hybridization. Cytogenet Genome Res 126 (4):368-375. doi:https://.doi.org/10.1159/000275796
McKain MR, McNeal JR, Kellar PR, Eguiarte LE, Pires JC, Leebens-Mack J (2016) Timing of rapid diversification and convergent origins of active pollination within Agavoideae (Asparagaceae). Am J Bot 103 (10):1717-1729. doi:https://.doi.org/10.3732/ajb.1600198
McKain MR, Wickett N, Zhang Y, Ayyampalayam S, McCombie WR, Chase MW, Pires JC, dePamphilis CW, Leebens-Mack J (2012) Phylogenomic Analysis of Transcriptome Data Elucidates Co-Occurrence of a Paleopolyploid Event and the Origin of Bimodal Karyotypes in Agavoideae (Asparagaceae). Am J Bot 99 (2):397-406. doi:https://.doi.org/10.3732/ajb.1100537
Pearce SR, Pich U, Harrison G, Flavell AJ, Heslop-Harrison JS, Schubert I, Kumar A (1996) The Ty1-copia group retrotransposons of Allium cepa are distributed throughout the chromosomes but are enriched in the terminal heterochromatin. Chromosome Res 4 (5):357-364. doi:https://.doi.org/10.1007/bf02257271
Qian H, Jin Y (2016) An updated megaphylogeny of plants, a tool for generating plant phylogenies and an analysis of phylogenetic community structure. J Plant Ecol 9 (2):233-239. doi:https:///10.1093/jpe/rtv047
Ramos LC, Baez M, Fuchs J, Houben A, Carvalho R, Pedrosa-Harand A (2023) Differential Repeat Accumulation in the Bimodal Karyotype of Agave L. Genes 14 (2). doi:https://.doi.org/10.3390/genes14020491
Rey MD, Moore G, Martín AC (2018) Identification and comparison of individual chromosomes of three accessions of Hordeum chilense, Hordeum vulgare, and Triticum aestivum by FISH. Genome 61 (6):387-396. doi:10.1139/gen-2018-0016
Robert ML, Lim KY, Hanson L, Sanchez-Teyer F, Bennett MD, Leitch AR, Leitch IJ (2008) Wild and agronomically important Agave species (Asparagaceae) show proportional increases in chromosome number, genome size, and genetic markers with increasing ploidy. Botanical Journal of the Linnean Society 158 (2):215-222. doi:https://.doi.org/10.1111/j.1095-8339.2008.00831.x
Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27 (6):207-216. doi:https://doi.org/10.1016/j.tig.2011.03.004
Sharma A, Presting GG (2008) Centromeric retrotransposon lineages predate the maize/rice divergence and differ in abundance and activity. Mol Genet Genomics 279 (2):133-147. doi:10.1007/s00438-007-0302-5
Shibata F, Nagaki K, Yokota E, Murata M (2013) Tobacco karyotyping by accurate centromere identification and novel repetitive DNA localization. Chromosome Res 21 (4):375-381. doi:https://.doi.org/10.1007/s10577-013-9363-y
Steele PR, Hertweck KL, Mayfield D, McKain MR, Leebens-Mack J, Pires JC (2012) Quality and quantity of data recovered from massively parallel sequencing: Examples in Asparagales and Poaceae. Am J Bot 99 (2):330-348. doi:https://doi.org/10.3732/ajb.1100491
Stull GW, Pham KK, Soltis PS, Soltis DE (2023) Deep reticulation: the long legacy of hybridization in vascular plant evolution. The Plant Journal 114 (4):743-766. doi:https://doi.org/10.1111/tpj.16142
Tank DC, Eastman JM, Pennell MW, Soltis PS, Soltis DE, Hinchliff CE, Brown JW, Sessa EB, Harmon LJ (2015) Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytol 207 (2):454-467. doi:https://doi.org/10.1111/nph.13491
Ullah I, Ansari HA, Verry IM, Hussain SW, Ellison NW, McManus MT, Williams WM (2022) Introgression of Trifolium ambiguum Into Allotetraploid White Clover (Trifolium repens) Using the Ancestral Parent Trifolium occidentale as a Bridging Species. Front Plant Sci 13. doi:https://.doi.org/10.3389/fpls.2022.858714
Vimala Y, Lavania S, Lavania UC (2021) Chromosome change and karyotype differentiation-implications in speciation and plant systematics. Nucleus Calcutta 64 (1):33-54. doi:https://.doi.org/10.1007/s13237-020-00343-y
Vondrak T, Robledillo LA, Novak P, Koblizkova A, Neumann P, Macas J (2020) Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant J 101 (2):484-500. doi:https://.doi.org/10.1111/tpj.14546
Wang Q, Wang Y, Wang JH, Gong Z, Han GZ (2021a) Plants acquired a major retrotransposon horizontally from fungi during the conquest of land. New Phytol. doi:https://10.1111/nph.17568
Wang YJ, Wang SW, Jia XJ, Tian ZR, Wang YF, Wang CY, Zhang H, Liu XL, Zhao JX, Deng PC, Ji WQ (2021b) Chromosome karyotype and stability of new synthetic hexaploid wheat. Mol Breeding 41 (10). doi:https://.doi.org/10.1007/s11032-021-01253-w
Weiss-Schneeweiss H, Schneeweiss GM (2013) Karyotype Diversity and Evolutionary Trends in Angiosperms. In: Greilhuber J, Dolezel J, Wendel JF (eds) Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Springer Vienna, Vienna, pp 209-230. doi:https://.doi.org/10.1007/978-3-7091-1160-4_13
Wu YX, Chen JH, He QL, Zhu SJ (2013) Parental origin and genomic evolution of tetraploid species by molecular marker and GISH analyses. Caryologia 66 (4):368-374. doi:https://.doi.org/10.1080/00087114.2013.857830
Yu-xiang W, Jin-hong C, Qiu-ling H, Shui-jin Z (2013) Parental origin and genomic evolution of tetraploid Gossypium species by molecular marker and GISH analyses. Caryologia 66 (4):368-374. doi:10.1080/00087114.2013.857830
Downloads
Published
License
Copyright (c) 2025 POLIBOTANICA

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.

Polibotánica by Departamento de Botánica de la Escuela Nacional de Ciencias Biológicas del Instituto Politécnico Nacional se distribuye bajo una Licencia Creative Commons Atribución-NoComercial-CompartirIgual 4.0 Internacional.



















